Molecular Mechanisms of Disease – Genes & Proteins
Genetic Mutations
Research into NF1 has tended to follow trends in molecular and biochemical techniques. With advancements in technology, mechanisms at the genetic, molecular, and cellular levels that NF1 utilizes for its pathogenesis have begun to become elucidated, providing greater insight into how the disease functions. Through all levels of our discussion of NF1, the trend of extreme variability will be present. This variability in onset of disease makes it difficult for biochemical characterization, and even treatment, of the disease.
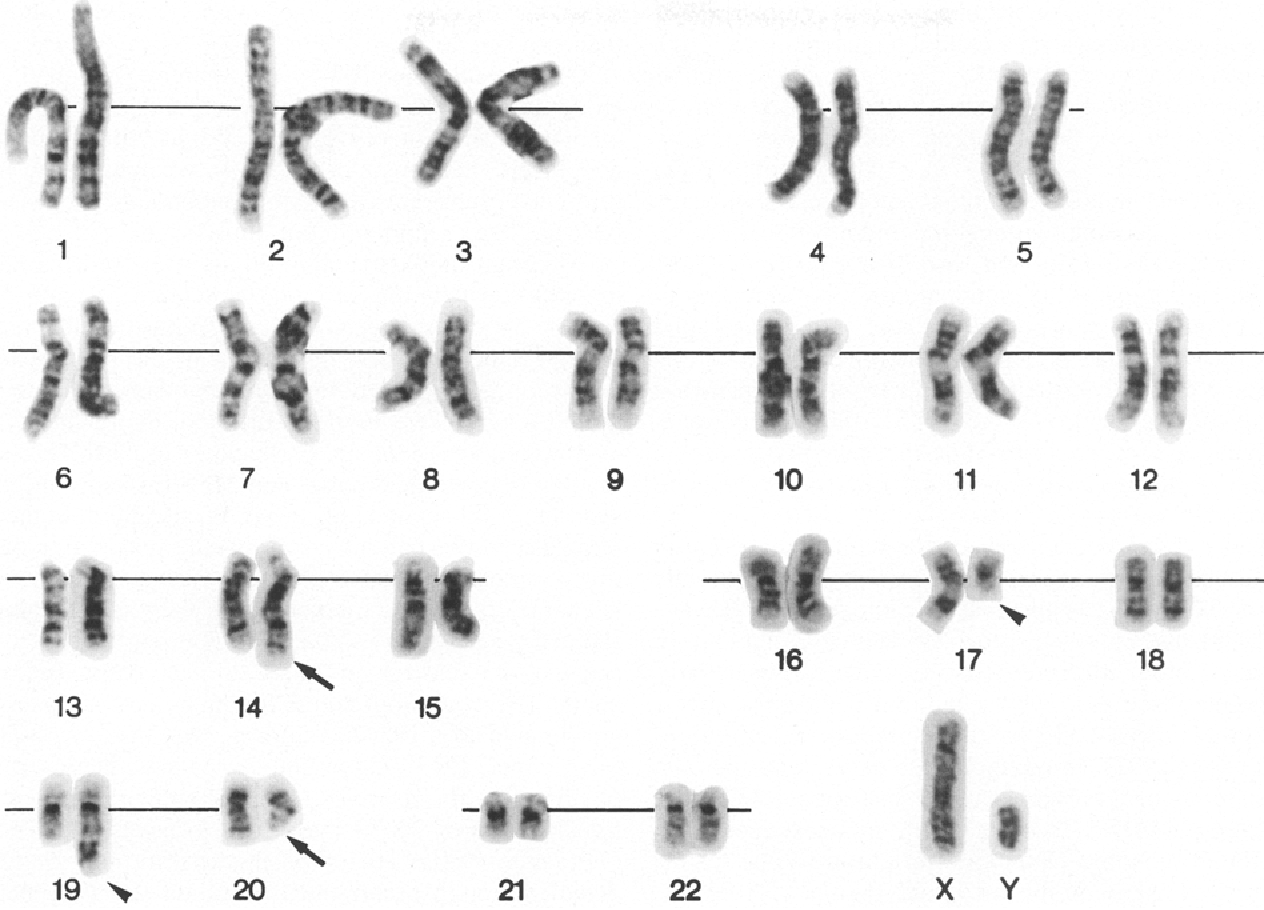
With the advent of recombinant DNA techniques in the 1970s, research into the genetic basis of NF1 quickly followed suit. Understanding the genetic mutations conferring NF1 will provide a good foundation for understanding the downstream effects of the disease. The story of the location of the genetic mutation leading to NF1 does not start with NF1, but neurofibromatosis type 2 (NF2). NF2 is more rare than NF1, and is characterized by tumors on the eighth cranial nerve.1 Extensive genetic studies were conducted by the Seizinger group on NF2 using polymorphic DNA markers to center its genetic mutation to chromosome 22.2,3 Using the same technique, Seizinger extended this work to NF1, and found that the NF1 mutation generally lies on the locus encoding the receptor for nerve growth factor on chromosome 17.3,4
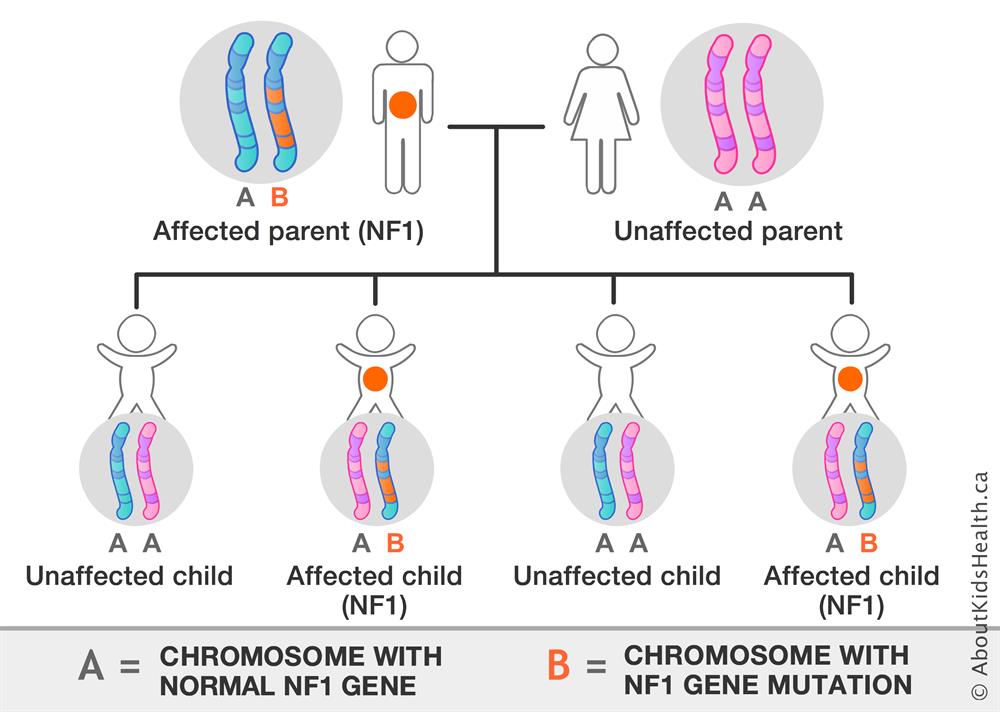
Allelic location of the NF1 mutation set the stage for deeper genotype work on the disease. Mutations have been found to occur near translocation breakpoints on chromosome 17.3,4 Specifically, a t(1;17) translocation or a t(17;22) translocation in the NF1 gene causes NF1 disease.3,4 This gene mutation is dominant and can be inherited from a parent or occur spontaneously (the number of cases inherited and acquired due to spontaneous mutation are roughly equal).5 As genotyping techniques improved, an increased number of genetic mutations leading to NF1 have been found. The genetic mutations are evenly distributed throughout the NF1 gene, and the number of them seems arbitrary to a certain extent (there are at least 33 publications on novel mutations leading to NF1). The mutations causing NF1 include ones leading to errors in mRNA splicing of the NF1 gene product, leading to prematurely truncated proteins being translated and mutations directly affecting the fragile structure of the protein, both leading to decreased functionality of the gene product.6 With the discovery of the location of mutations causing NF1, Xu et al. were able to utilize cDNA technology to find that the product of the mutated gene has significant homology to the catalytic domain of the yeast IRA1 product, which was known at the time to be very similar to the mammalian GTPase-activating protein (GAP).3 The GTPase activity of the NF1 gene product, neurofibromin, was confirmed in the same year as it was discovered that it was able to change the phosphorylation state of ras p21.7
Protein Mutations
Concurrently with the discovery of the translocation causing NF1, it was found that neurofibromin has significant homology to the yeast IRA1 product, which is very similar to the mammalian GTPase-activating protein (GAP).3 The importance of the mammalian GAP to cell-signaling pathways led to hypotheses heralding the role of neurofibromin to be that of regulating cell-signaling pathways. Later studies found that neurofibromin stimulates GTPase activity of ras p21, with a much higher affinity than mammalian GAP, confirming the GAP activity of neurofibromin.7 Although the mutations leading to NF1 are numerous and may vary significantly from one another, general trends can be understood at the level of the neurofibromin protein. The neurofibromin crystal structure was only solved in 2021, but this breakthrough allowed for illuminating insight into the molecular scaffolding governing NF1 disease. Neurofibromin must assemble into a homodimer to be functional. The quaternary structure of the protein takes on a “figure-eight”-like shape. The “core” of the protein, required for assembly of the homodimer, is formed by the N- and C-HEAT domains of each protein comprising the dimer coming together, in a “helical packing” interface.8 Mutations have been found affecting both the N-HEAT (L844F) and C-HEAT (L1834R, N1840K, L2104R) domains, interfering with helical packing of the homodimer, leading to neurofibromin being ineffective and causing NF1.8 Other mutations have been associated with destabilizing the neurofibromin dimer interface.8 The variety and seemingly uniform spread of these mutations suggest that the complexity of the folds of the neurofibromin protein make it so that any mutations are likely to lead to disease, regardless of location.18 This suggestion is strengthened when the primary sequence of human neurofibromin is compared across different species. Neurofibromin has been found to be highly conserved in mammals, with the human neurofibromin primary sequence being 98.5% to that found in rats, and 99.4% to that found in dogs, implying strong selective pressures on the structure and function of neurofibromin.7 Nature tends to conserve the structure of key proteins and enzymes. This significant conservation in sequence implies the importance of the specific amino acids that comprise neurofibromin, elucidating the adverse effects that point mutations can have.
Neurofibromin is a regulatory protein that is very susceptible to loss-of-function (LOF) mutations. Nature may have evolved single regulatory proteins controlling a multitude of pathways because of the efficiency and energy conservation this grants. However, when one protein is responsible for the operation of multiple crucial signaling pathways, there is a cause of worry for mutations. The susceptibility of neurofibromin to mutations is intriguing due to its presence in many mammals, and its importance for survival. Nevertheless, neurofibromatosis type I is a disease that starts at the genetic level, where mutations (either inherited or acquired) on chromosome 17 have effects on neurofibromin. The severity of this disease is due to the importance of neurofibromin, and its role in regulating cytosolic cell pathways.
Works Cited
For a complete list of works used, please access the Annotated Bibliography.
3 Comments
Ethan Forrer · May 6, 2022 at 8:06 pm
Great job thoroughly exploring and describing a disease that affects such a core system in cell signalling. It’s not surprising that issues with such a fundamental system could lead to such a range of symptoms. You mentioned that number of cases of spontaneously acquired NF1 are roughly equal to inherited cases, which is unusual considering it is such a relatively well conserved gene and a dominate mutation with a relatively low mortality rate. Have you found any sources that described a relationship between severity of symptoms of NF1 in inherited vs. spontaneously acquired patients?
Dave Strzeminski · May 9, 2022 at 11:30 pm
Hi Mike. Great work! I was wondering if once the mutations occur in neurofibromin and you display symptoms of NF1, if there are any therapeutic options for someone afflicted with it, and if there is, could you show me a structure of the drug? I’m curious to see how it binds.
admin · May 12, 2022 at 2:48 pm
Hey, Dave! Thank you so much for this question! There has been somewhat of a breakthrough treatment, although not wholly approved for all people afflicted with NF1. It is a drug named selumetinib, and to understand how it works, it is important to remember what causes NF1. NF1 occurs when a regulatory protein, neurofibromin, is mutated and no longer functional. Neurofibromin is a GTPase, responsible for turning off pathways leading to cellular proliferation. Without neurofibromin, this pathway is able to go on, inhibited, leading to the uncontrolled growth of cells, i.e. tumors. Selumetinib is a selective inhibitor of MEK 1 and 2, downstream signaling molecules in this proliferation pathway. By inhibiting these downstream messengers, selumetinib is able to inhibit the pathway, and restrict uncontrolled cell division. Currently, it is only approved for use in children with severe tumors. Other than that, the most viable option for “treatment” is the removal of tumors that show signs of becoming malignant, restrict someone from performing daily tasks, or that cause severe pain. Here is a link to the structure of the drug (https://pubchem.ncbi.nlm.nih.gov/compound/Selumetinib#:~:text=Selumetinib%20is%20a%20member%20of,role%20as%20an%20EC%202.7.), maybe you do some research on this in grad school!